Key Features¶
Data Conversion¶
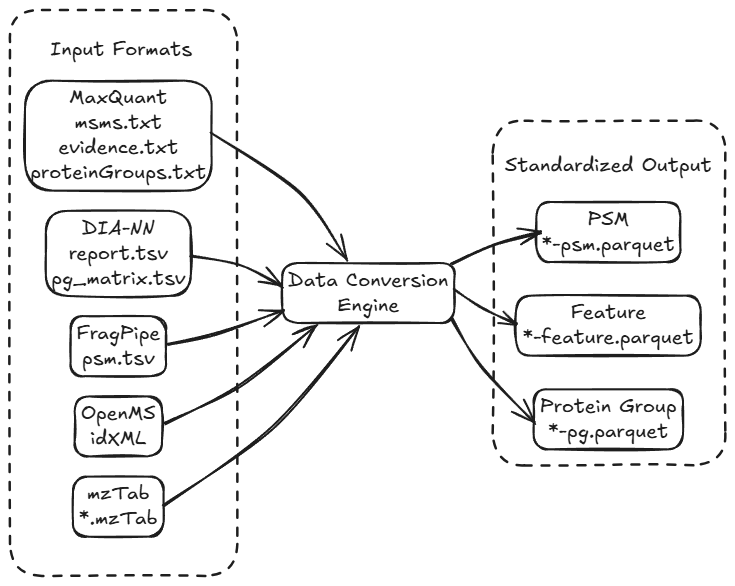
Convert data from leading proteomics software to standardized QPX format:
- MaxQuant result files:
msms.txt: MS/MS scan information (PSM data)evidence.txt: Peptide evidence (Feature data)proteinGroups.txt: Protein identification results-
*.sdrf.tsv: SDRF-Proteomics metadata (optional) -
DIA-NN result files:
report.tsvorreport.parquet: DIA-NN main reportpg_matrix.tsv: Protein group matrix*.sdrf.tsv: SDRF-Proteomics metadata (optional)-
*ms_info.parquet: mzML statistics folder (optional) -
FragPipe result files:
psm.tsv: PSM identifications-
*.sdrf.tsv: SDRF-Proteomics metadata (optional) -
OpenMS/idXML files:
*.idXML: Identification results-
*.mzML: Corresponding spectra files (optional, for batch processing) -
mzTab (quantms pipeline) files:
*.mzTab: Standard proteomics format (PSM/PEP/PRT sections)*msstats*.csv: MSstats/MSstatsTMT input files (optional)
Output format: All conversions produce standardized parquet files:
- PSM:
*-psm.parquet - Feature:
*-feature.parquet - Protein Group:
*-pg.parquet
Data Transformation¶
Transform and enrich your data:
- Absolute Expression (AE): iBAQ-based absolute quantification
- Differential Expression (DE): MSstats statistical analysis
- Gene Mapping: Add gene annotations
- UniProt Integration: Latest protein annotations
- AnnData Export: Integration with single-cell tools
Visualization¶
Generate publication-quality plots:
- Intensity distributions (KDE, box plots)
- Peptide distribution analysis
- iBAQ distribution plots
- Quality control visualizations
Statistical Analysis¶
Comprehensive data statistics:
- PSM data statistics (proteins, peptides, PSMs count)
- Project-level absolute expression statistics
- Sample-level quantitative metrics
- Automated report generation
Project Management¶
Track and share your work:
- PRIDE Archive integration
- Project metadata management
- File registration and tracking